Protein structure
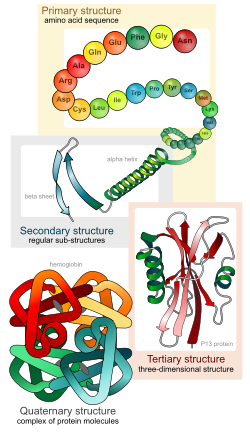

Protein structure describes how protein molecules are organised. This structure is what makes proteins work.
Proteins are important biological macromolecules present in all organisms. They are polymers formed from 20 possible amino acids by RNA translation. Protein structures range in size from tens to several thousand amino acids.[1]
After translation, proteins fold into specific shapes. This is not done by chemical bonds but by weaker forces such as hydrogen bonds. To understand how proteins work, it is often necessary to discover their three-dimensional structure. To do this biophysics uses techniques such as X-ray crystallography, NMR spectroscopy, and dual polarisation interferometry.
A protein may switch from one shape to another as it does its job. The alternative states of the same protein are called conformations. An enzyme, for instance, will have at least two conformations: one with its co-enzyme and one without. The form with its coenzyme will have two conformations: one with its substrate and one without.
Levels of protein structure
[change | change source]There are four distinct levels of protein structure.
Primary structure
[change | change source]Proteins are molecules that consist of one or more chains of amino acids. Each different protein has a set of amino acids in a particular order. The primary structure is the sequence of the amino acids of a single polypeptide chain from its start (N-terminus) to its end (C-terminus). The amino acids are held together by a specific type of covalent bond, known as a peptide bond, which are made during the process of protein biosynthesis or translation.
Secondary structure
[change | change source]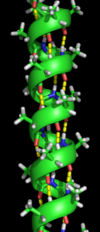
Secondary structure is the most simple structure made by proteins' polypeptide chains. There are two main types of secondary structure: alpha helix and beta strand (beta sheet). Alpha helices are coils that turn in a clockwise rotation. Beta strands are chains of amino acids that lie beside each other to make a beta sheet. These two types of secondary structure were suggested in 1951 by Linus Pauling and coworkers.[2] Unlike the polypeptide chain, which is bonded through covalent peptide bonds, secondary structure is formed by hydrogen bonds.
Tertiary structure
[change | change source]This is the shape (spatial organization) of an entire protein molecule. Protein folding is largely self-organising. It is mainly decided by the protein's primary structure.[3] This is called Anfinsen's dogma.[4] However, the environment in which a protein is made and folded also effect its final shape.
Quaternary structure
[change | change source]If proteins are built of more than one polypeptide, this gives another level of structure. The quaternary structure, is how the polypeptide subunits fit together. Haemoglobin, for example, has two alpha and two beta chains. Both the alpha chains and beta chains are coded for by a cluster of six or seven genes, which provide the code for different versions of the molecule.[5][6]
References
[change | change source]- ↑ Brocchieri L, Karlin S (2005-06-10). "Protein length in eukaryotic and prokaryotic proteomes". Nucleic Acids Research. 33 (10): 3390–3400. doi:10.1093/nar/gki615. PMC 1150220. PMID 15951512.
- ↑ Pauling L, Corey RB, Branson HR (1951). "The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain". Proc Natl Acad Sci USA. 37 (4): 205–211. Bibcode:1951PNAS...37..205P. doi:10.1073/pnas.37.4.205. PMC 1063337. PMID 14816373.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Alberts, Bruce et al 2002. The shape and structure of proteins. In Molecular biology of the cell. Now in its 7th edition . New York and London: Garland Science. ISBN 0-8153-3218-1
- ↑ Anfinsen C. 1972. The formation and stabilization of protein structure. Biochem. J. 128 (4): 737–49. [1]
- ↑ Higgs D.R.; et al. (1989). "A review of the molecular genetics of the human alpha-globin gene cluster". Blood. 73 (5): 1081–104. doi:10.1182/blood.V73.5.1081.1081. PMID 2649166.
- ↑ Levings PP, Bungert J (2002). "The human beta-globin locus control region". Eur. J. Biochem. 269 (6): 1589–99. doi:10.1046/j.1432-1327.2002.02797.x. PMID 11895428.
Other websites
[change | change source]- Education portal: Proteins IV: primary, secondary, tertiary and quaternary structure. [2]
