Virus
| Virus | |
|---|---|
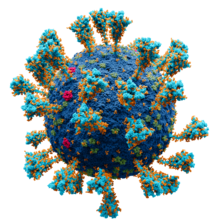
| |
| SARS-CoV-2, a member of the subfamily Coronavirinae | |
| Virus classification | |
| (unranked): | Virus |
| Realms | |

A virus is a tiny parasite.[1] Virology is the study of viruses.
Viruses can only be seen under an electron microscope. Viruses are not free-living: they can only be parasites. They always reproduce inside other living things. All viruses infect living organisms, and may cause disease. The virus make copies of itself inside another organism's cells. Viruses are a strand of nucleic acid with a protein coat. Usually the nucleic acid is RNA; sometimes it is DNA. Viruses cause many types of diseases, such as polio, ebola and hepatitis.
Viruses reproduce by getting their nucleic acid strand into a prokaryote or eukaryote cell. The RNA or DNA strand then takes over the cell machinery to reproduce copies of itself and the protein coat. The cell then bursts open, spreading the newly created viruses. All viruses reproduce this way, and there are no free-living viruses.[2][3] Viruses are everywhere in the environment, and all organisms can be infected by them.[4][5][6]
Most viruses are much smaller than bacteria. They were not visible until the invention of the electron microscope. A virus has a simple structure. It has just a protein coat which covers a string of nucleic acid. Viruses live and reproduce inside the cells of other living organisms.
With eukaryotic cells, the virus protein coat is able to enter the target cells by certain cell membrane receptors. With prokaryote bacteria cells, the bacteriophage physically injects the nucleic acid strand into the host cell.
Viruses have the following characteristics:
- They outnumber all other forms of life on the planet by a long way.[1]
- They are infectious particles, causing many types of disease;
- They contain a nucleic acid core of RNA or DNA;
- They are surrounded by a protective protein coat;
When the host cell has finished making more viruses, it undergoes lysis, or breaks apart. The viruses are released and are then able to infect other cells. Viruses can remain "silent" (inactive) for a long time, and will infect cells when the time and conditions are right.
Some special viruses are worth noting. Bacteriophages have evolved to enter bacterial cells, which have a different type of cell wall from eukaryote cell membranes. Envelope viruses, when they reproduce, cover themselves with a modified form of the host cell membrane, thus gaining an outer lipid layer that helps entry. Some of our most difficult-to-combat viruses, like influenza and HIV, use this method.
Viral infections in animals trigger an immune response which usually kills the infecting virus. Vaccines can also produce immune responses. They give an artificially acquired immunity to the specific viral infection. However, some viruses (including those causing AIDS and viral hepatitis) escape from these immune responses and cause chronic infections. Antibiotics have no effect on viruses, but there are some other drugs which can be used against viruses.
Genome[change | change source]
| Property | Parameters |
|---|---|
| Nucleic acid |
|
| Shape |
|
| Strandedness |
|
| Sense |
|

There are many genomic structures in viruses. As a group they are more diverse than plants, animals, archaea, or bacteria. There are millions of different types of viruses,[4] but only about 7,000 of them have been described in detail.[2]49
A virus has either RNA or DNA genes and so is called an RNA virus or a DNA virus. The vast majority of viruses have RNA genomes. Plant viruses usually have single-stranded RNA genomes and bacteriophages usually have double-stranded DNA genomes.[7]96/99
Fewer than 7,000 types have been described in detail. but there are doubtlessly many more to be discovered.[8][9]
Replication cycle[change | change source]
Viral populations do not grow through cell division, because they do not have cells. Instead, they use the machinery and metabolism of a host cell to produce many copies of themselves, and they assemble (put together) in the cell.
The life cycle of viruses differs greatly between species but there are six basic stages in the life cycle of viruses:[7]75/91
- Attachment is a binding between viral capsid proteins and specific receptors on the host cellular surface.
- Penetration follows attachment: Virions (single virus particles) enter the host cell by receptor-mediated endocytosis or fusion with the lipid bilayer. This is called viral entry.
The infection of plant and fungal cells is different from that of animal cells. Plants have a rigid cell wall made of cellulose, and fungi one of chitin. This means most viruses can only get inside these cells by force.[2]70 An example would be: a virus travels on an insect vector which feeds on plant sap. The damage done to cell walls would let the virus get in.
Bacteria, like plants, have strong cell walls that a virus must get through to infect the cell. However, bacterial cell walls are much thinner than plant cell walls, and some viruses have mechanisms that inject their genome into the bacterial cell, while the viral capsid remains outside.[2]71 - Uncoating is how the viral capsid is removed: This may be by viral enzymes or host enzymes or by simple dissociation; the end-result is the releasing of the viral nucleic acid.
- Replication of viruses is multiplying the genome. This usually means production of viral messenger RNA (mRNA) from "early" genes. This may be followed, for complex viruses, by one or more further rounds of mRNA synthesis: "late" gene expression is of structural or virion proteins.
- After the self-assembly of the virus particles, some modification of the proteins often occurs. In viruses such as HIV, this modification (sometimes called maturation) occurs after the virus has been released from the host cell.[10]
- Viruses can be released from the host cell by lysis, a process that kills the cell by bursting its membrane and cell wall. This is a feature of many bacterial and some animal viruses.
In some viruses the viral genome is put by genetic recombination into a specific place in the host's chromosome. The viral genome is then known as a "provirus" or, in the case of bacteriophages a "prophage".[3]60
Whenever the host divides, the viral genome is also replicated. The viral genome is mostly silent within the host; however, at some point, the provirus or prophage may give rise to active virus, which may lyse the host cells.[2]chapter 15
Enveloped viruses (e.g. HIV) typically are released from the host cell after the virus acquires its envelope. The envelope is a modified piece of the host's plasma membrane.[2]185/7
Genetic material and replication[change | change source]
The genetic material within virus particles, and the method by which the material is replicated, varies considerably between different types of viruses.
- RNA viruses
- Replication usually takes place in the cytoplasm. RNA viruses can be placed into four different groups depending on their modes of replication. All RNA viruses use their own RNA replicase enzymes to create copies of their genomes.[7]79
- DNA viruses
- The genome replication of most DNA viruses takes place in the cell's nucleus. Most DNA viruses are entirely dependent on the host cell's DNA and RNA synthesising machinery, and RNA processing machinery. Viruses with larger genomes may encode much of this machinery themselves. In eukaryotes the viral genome must cross the cell's nuclear membrane to access this machinery, while in bacteria it need only enter the cell.[3]54[7]78
- Reverse transcribing viruses
- Reverse transcribing viruses with RNA genomes (retroviruses) use a DNA intermediate to replicate. Those with DNA genomes (pararetroviruses) use an RNA intermediate during genome replication.[11] They are susceptible to antiviral drugs that inhibit the reverse transcriptase enzyme. An example of the first type is HIV, which is a retrovirus. Examples of the second type are the Hepadnaviridae, which includes Hepatitis B virus.[7]88/9
Host defence mechanisms[change | change source]
Innate immune system[change | change source]
The body's first line of defence against viruses is the innate immune system. This has cells and other mechanisms which defend the host from any infection. The cells of the innate system recognise, and respond to, pathogens in a general way.[12]
RNA interference is an important innate defence against viruses.[13] Many viruses have a replication strategy that involves double-stranded RNA (dsRNA). When such a virus infects a cell, it releases its RNA molecule. A protein complex called dicer sticks to it and chops the RNA into pieces. Then a biochemical pathway, called the RISC complex, starts up. This attacks the viral mRNA, and the cell survives the infection.
Rotaviruses avoid this by not uncoating fully inside the cell and by releasing newly produced mRNA through pores in the particle's inner capsid. The genomic dsRNA remains protected inside the core of the virion.[14][15]
The production of interferon is an important host defence mechanism. This is a hormone produced by the body when viruses are present. Its role in immunity is complex; it eventually stops the viruses from reproducing by killing the infected cell and its close neighbours.[16]
Adaptive immune system[change | change source]
Vertebrates have a second, more specific, immune system. It is called the adaptive immune system. When it meets a virus, it produces specific antibodies that bind to the virus and render it non-infectious. Two types of antibodies are important.
The first, called IgM, is highly effective at neutralizing viruses but is produced by the cells of the immune system only for a few weeks. The second, called IgG, is produced indefinitely. The presence of IgM in the blood of the host is used to test for acute infection, whereas IgG indicates an infection sometime in the past.[17] IgG antibody is measured when tests for immunity are carried out.[18]
Another vertebrate defence against viruses involves immune cells known as T cells. The body's cells constantly display short fragments of their proteins on the cell's surface, and, if a T cell recognises a suspicious viral fragment there, the host cell is destroyed by killer T cells and the virus-specific T-cells proliferate. Cells such as macrophages are specialists at this antigen presentation.[19]
Evading the immune system[change | change source]
Not all virus infections produce a protective immune response. These persistent viruses evade immune control by sequestration (hiding away); cytokine resistance; evading natural killer cell activity; escape from apoptosis (cell death), and antigenic shift (changing surface proteins).[20] HIV evades the immune system by constantly changing the amino acid sequence of the proteins on the surface of the virion. Other viruses move along nerves to places the immune system cannot reach.
Evolution[change | change source]
Viruses do not belong to any of the six kingdoms. They do not meet all the requirements for being classified as a living organism because they are not active until the point of infection. However, that is just a verbal point.
Obviously, their structure and mode of operation means they have evolved from other living things, and the loss of normal structure occurs in many endoparasites. The origins of viruses in the evolutionary history of life are unclear: some may have evolved from plasmids – pieces of DNA that can move between cells – while others may have evolved from bacteria. In evolution, viruses are an important means of horizontal gene transfer, which increases genetic diversity.[21]
Recent discoveries[change | change source]
A recent project discovered nearly 1500 new RNA viruses by sampling over 200 invertebrate species. "The research team... extracted their RNA and, using next-generation sequencing, deciphered the sequence of a staggering six trillion letters present in the invertebrate RNA libraries".[22] The research showed that viruses changed bits and pieces of their RNA by a variety of genetic mechanisms. "The invertebrate virome [shows] remarkable genomic flexibility that includes frequent recombination, lateral gene transfer among viruses and hosts, gene gain and loss, and complex genomic rearrangements".[23]
How bacteria and archaea deal with viruses[change | change source]
Viruses have been on this planet a long time. We now know that bacteria and archaea had to deal with them first, before our type of cellular life evolved. Details of the defence mechanisms used by archaea and bacteria are discussed on the page CRISPR, which briefly introduces the topic of early defences against viruses.
Largest virus[change | change source]
A group of large viruses infect amoebae. The largest is Pithovirus. Others in order of size are Pandoravirus, then Megavirus, then Mimivirus. They are bigger than some bacteria, and visible under a light microscope.
Viruses in the sea[change | change source]
Viruses are everywhere in the sea. They may outnumber all other forms of marine life by at least an order of magnitude. Through selective infection, viruses influence nutrient cycling and evolution in the ocean.[24]
Uses[change | change source]
Viruses are used widely in cell biology.[25] Geneticists often use viruses as vectors to introduce genes into cells that they are studying. This is useful for making the cell produce a foreign substance, or to study the effect of introducing a new gene into the genome. Eastern European scientists have used phage therapy as an alternative to antibiotics for some time, and interest in this approach is increasing, because of the high level of antibiotic resistance now found in some pathogenic bacteria.[26]
Related pages[change | change source]
References[change | change source]
- ↑ 1.0 1.1 Glaunsinger, Britt 2020 Viruses reveal the secrets of biology. [1] This source makes the astonishing claim that there are !024 viral infections/second.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Dimmock N.J; Easton, Andrew J; Leppard, Keith 2007. Introduction to modern virology. 6th ed, Blackwell. ISBN 1-4051-3645-6
- ↑ 3.0 3.1 3.2 Shors Teri 2017. Understanding viruses. Jones and Bartlett. ISBN 978-1-284-02592-7
- ↑ 4.0 4.1 Breitbart M, Rohwer F (2005). "Here a virus, there a virus, everywhere the same virus?". Trends Microbiol. 13 (6): 278–84. doi:10.1016/j.tim.2005.04.003. PMID 15936660.
- ↑ Lawrence CM, Menon S, Eilers BJ; et al. (2009). "Structural and functional studies of archaeal viruses". J. Biol. Chem. 284 (19): 12599–603. doi:10.1074/jbc.R800078200. PMC 2675988. PMID 19158076.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Edwards RA, Rohwer F (2005). "Viral metagenomics". Nat. Rev. Microbiol. 3 (6): 504–10. doi:10.1038/nrmicro1163. PMID 15886693. S2CID 8059643.
- ↑ 7.0 7.1 7.2 7.3 7.4 Collier, Leslie; Balows, Albert; Sussman, Max 1998. Topley and Wilson's Microbiology and microbial infections. 9th ed, vol 1, Virology. volume editors: Mahy, Brian and Collier, Leslie. Arnold. ISBN 0-340-66316-2
- ↑ Pennisi E (March 2011). "Microbiology. Going viral: exploring the role of viruses in our bodies". Science. 331 (6024): 1513. Bibcode:2011Sci...331.1513P. doi:10.1126/science.331.6024.1513. PMID 21436418.
- ↑ Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, et al. (December 2016). "Redefining the invertebrate RNA virosphere". Nature. 540 (7634): 539–43. Bibcode:2016Natur.540..539S. doi:10.1038/nature20167. PMID 27880757. S2CID 1198891.
- ↑ Barman S, Ali A, Hui EK, Adhikary L, Nayak DP (2001). "Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses". Virus Res. 77 (1): 61–9. doi:10.1016/S0168-1702(01)00266-0. PMID 11451488.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Staginnus C, Richert-Pöggeler KR (2006). "Endogenous pararetroviruses: two-faced travelers in the plant genome". Trends in Plant Science. 11 (10): 485–91. doi:10.1016/j.tplants.2006.08.008. PMID 16949329.
- ↑ Alberts, Bruce; et al. (2002). Molecular biology of the cell; 4th ed. New York and London: Garland Science. ISBN 0-8153-3218-1. Retrieved 15 September 2008.
- ↑ Ding SW, Voinnet O (2007). "Antiviral immunity directed by small RNAs". Cell. 130 (3): 413–26. doi:10.1016/j.cell.2007.07.039. PMC 2703654. PMID 17693253.
- ↑ Patton JT, Vasquez-Del Carpio R, Spencer E (2004). "Replication and transcription of the rotavirus genome". Curr. Pharm. Des. 10 (30): 3769–77. doi:10.2174/1381612043382620. PMID 15579070.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Jayaram H, Estes MK, Prasad BV (2004). "Emerging themes in rotavirus cell entry, genome organization, transcription and replication". Virus Res. 101 (1): 67–81. doi:10.1016/j.virusres.2003.12.007. PMID 15010218.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Le Page C, Génin P, Baines MG, Hiscott J (2000). "Interferon activation and innate immunity". Rev Immunogenet. 2 (3): 374–86. PMID 11256746.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Greer S, Alexander GJ (1995). "Viral serology and detection". Baillieres Clin. Gastroenterol. 9 (4): 689–721. doi:10.1016/0950-3528(95)90057-8. PMID 8903801.
- ↑ Matter L, Kogelschatz K, Germann D (1997). "Serum levels of rubella virus antibodies indicating immunity: response to vaccination of subjects with low or undetectable antibody concentrations". J. Infect. Dis. 175 (4): 749–55. doi:10.1086/513967. PMID 9086126. S2CID 27281805.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Cascalho M, & Platt J.L (2007). "Novel functions of B cells". Crit. Rev. Immunol. 27 (2): 141–51. doi:10.1615/critrevimmunol.v27.i2.20. PMID 17725500.
- ↑ Hilleman M.R. (2004). "Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections". Proc. Natl. Acad. Sci. U.S.A. 101 (Suppl 2): 14560–6. Bibcode:2004PNAS..10114560H. doi:10.1073/pnas.0404758101. PMC 521982. PMID 15297608.
- ↑ Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6(4):417–24. doi:10.1016/S1369-5274(03)00086-9. PMID 12941415.
- ↑ Ball, Jonathan 2016. Bumper load of new viruses identified. BBC News Science & Environment. [2]
- ↑ Mang Shi et al 2016. Redefining the invertebrate RNA virosphere. Nature preview [3]
- ↑ Lang A.S; Rise L.R. Culley A.I. & Steward G.F. 2009. RNA viruses in the sea. [4]
- ↑ Lodish, Harvey; et al. 2008. Viruses: structure, function, and uses. [5] Retrieved 16 September 2008
- ↑ Matsuzaki S.; et al. (2005). "Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases". J. Infect. Chemother. 11 (5): 211–9. doi:10.1007/s10156-005-0408-9. PMID 16258815. S2CID 8107934.
